xGen™ Exome Hybridization Panel
Higher on-target percentage—even with a 12-plex capture
The xGen Exome Hyb Panel v2 is designed with a target-aware algorithm that minimizes off-target binding while maximizing coverage of coding regions. When combined with the xGen Exome Hyb Spike-in Panel v2.5, the result is best-in-class coverage across key databases, helping you confidently capture the most critical genomic targets.
xGen NGS—made for whole exome sequencing.
Ordering
Key Benefits of xGen Exome Hyb Panels
The xGen Exome Hyb Panel v2 includes 415,115 individually synthesized 5′ biotinylated probes, and the Spike-in Panel v2.5 adds 50,460 additional probes—each quality-tested and pooled at precise concentrations for consistent performance.
- Individually synthesized probes provide accurate representation and consistent quality across batches
- Higher on-target rate compared to leading array-based competitors due to probe-level quality control
Expanded coverage when combining both panels, capturing critical targets across:
- RefSeq 38.p6
- Ensembl
- CCDS
- HGMD
- COSMIC Cancer Gene Census
- ACMG v3.2
- ClinVar
This combination delivers comprehensive coverage for exome discovery research, ensuring your sequencing captures the most relevant genomic regions.
For customers who wish to purchase the legacy xGen™ Exome Hyb Panel v1, please contact us today.
For the xGen Hybridization and Wash Kit, xGen Universal Blockers, xGen Library Amplification Primer Mix, and/or xGen Human Cot DNA, please visit the xGen Hybridization Capture Core Reagents page.
Product details
xGen Exome Hyb Panel v2: Precision Targeting for Protein-Coding Regions
Exome sequencing focuses on the protein-coding regions of the genome—known as exons—which make up just 1% of the human genome. These regions are critical for identifying mutations that may contribute to disease. To study these mutations effectively, it's essential to isolate coding regions from noncoding DNA and sequence them at a depth sufficient for detecting variant alleles and copy number variations (CNVs).
The xGen Exome Hyb Panel v2 is designed to meet these needs by targeting only the coding sequences (CDS) of human genes listed in the RefSeq 109 database. This enables high multiplexing and deep coverage.
Engineered for Accuracy and Consistency
Each probe in the xGen Exome Hyb Panel v2 is a 5′ biotin–modified oligonucleotide, individually synthesized and verified using electrospray ionization-mass spectrometry (ESI-MS) and optical density (OD) measurements. Probes are normalized before pooling to ensure accurate representation, and any that fail quality control are resynthesized. This process establishes a level of quality and consistency that array-derived probe pools cannot match, where missing or truncated probes often go undetected.
Thanks to IDT’s proprietary synthesis methods, even probes with high GC or AT content are reliably represented. Probes are synthesized at a larger scale than typical array manufacturing, allowing for the creation of a single, large lot that is aliquoted over time—providing consistent performance across experiments.
Comprehensive Coverage with Advanced Design
The panel includes 415,115 probes spanning a 34 Mb target region across 19,435 human genes, with 39 Mb of probe space. Probe design is guided by a proprietary “capture-aware” algorithm and evaluated using off-target analysis to provide specificity and performance. All probes are manufactured under ISO 13485 standards, with dual quantification and mass spectrometry validation prior to pooling.
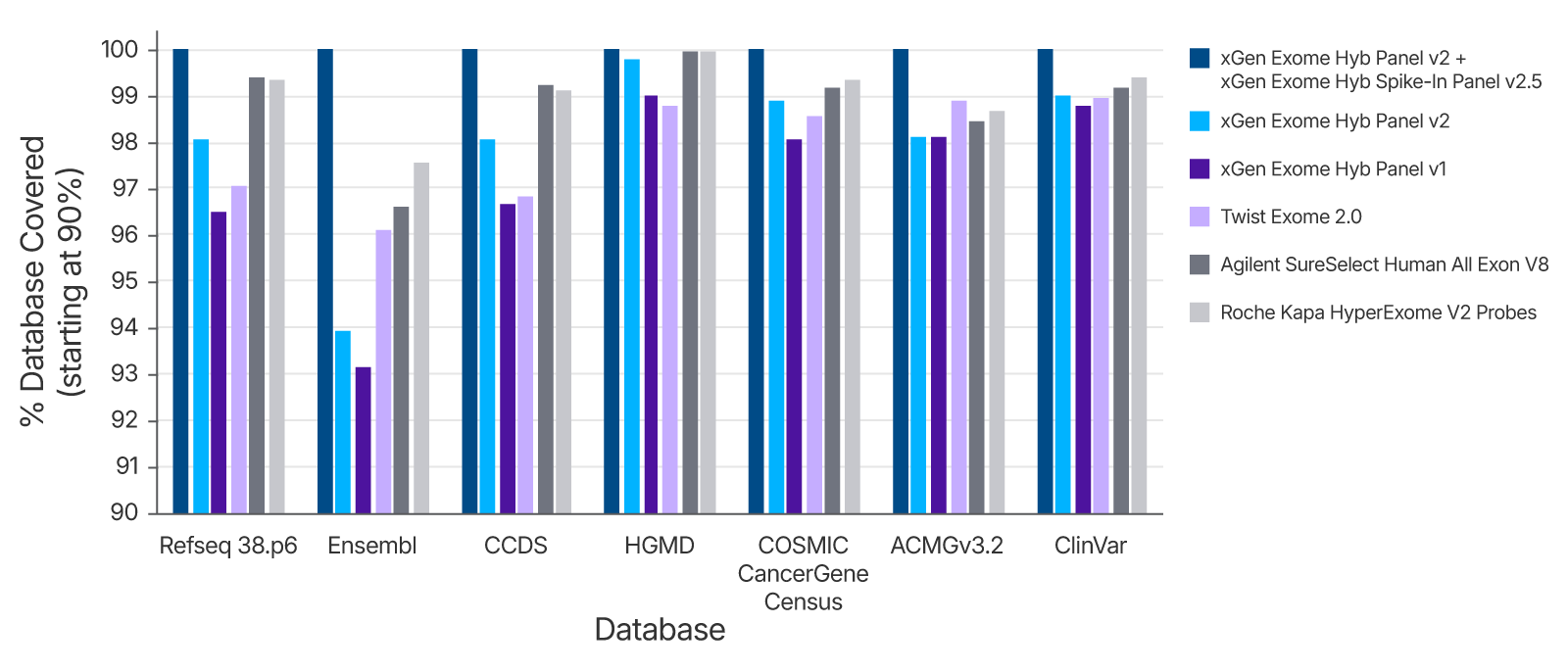
Expand Target Reach with Spike-in Panel v2.5
For researchers seeking broader coverage, combining the xGen Exome Hyb Panel v2 with the xGen Exome Hyb Spike-in Panel v2.5 unlocks access to newly identified targets from key databases, including:
- RefSeq 38.p6
- Ensembl
- CCDS
- HGMD
- COSMIC Cancer Gene Census
- ACMG v3.2
- ClinVar
This combination your sequencing efforts remain aligned with the latest genomic discoveries.
Automate Your NGS Workflow
Looking to improve lab efficiency and consistency? Explore our NGS Automation solutions to streamline your workflow and reduce manual steps—without compromising data quality.
Product data
Consistent data from one aliquot to the next
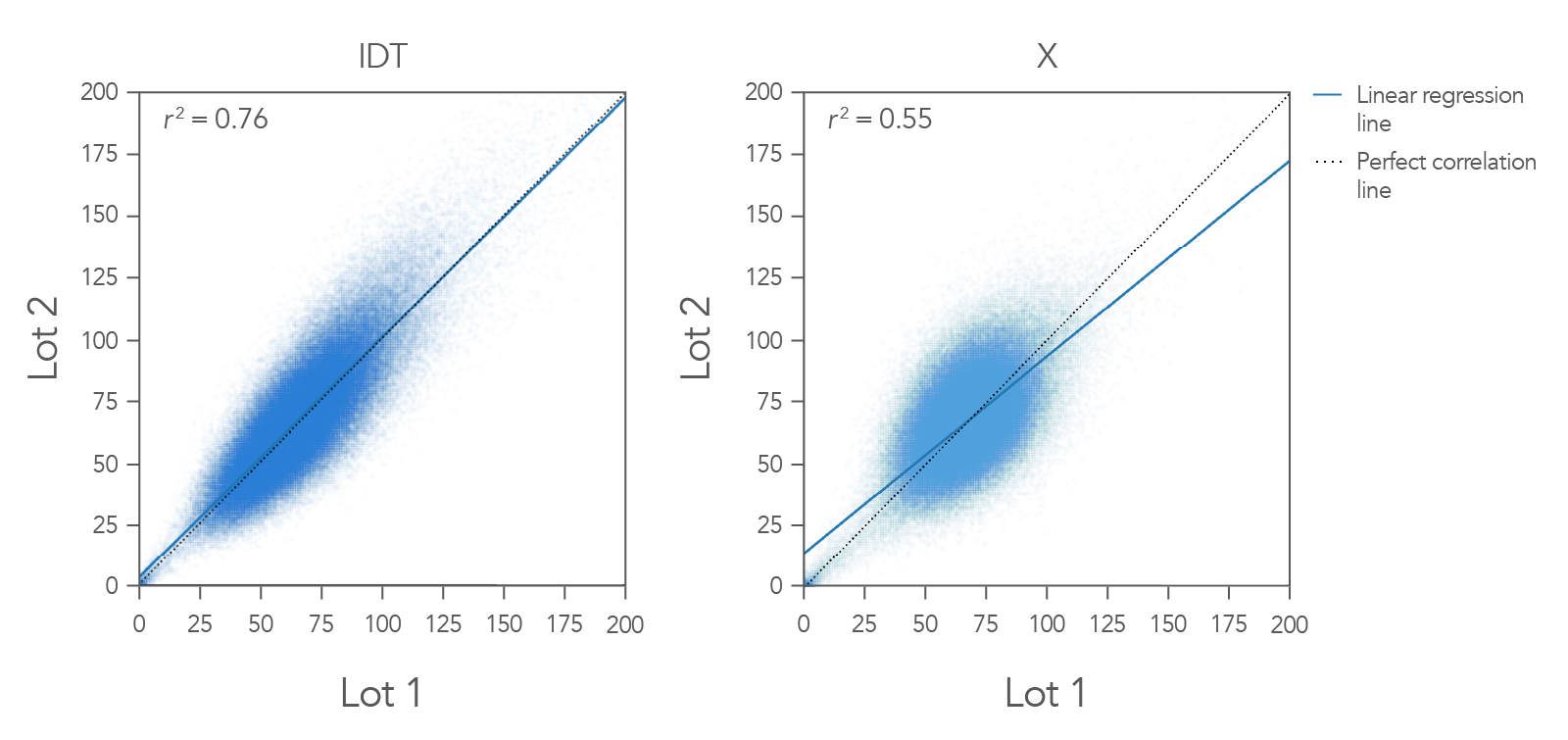
Figure 1. Example coverage of the IDT xGen Exome Hyb Panel v2 vs. competitor. Two different users performed hyb captures on different days in different locations using different aliquots (lots) of the IDT xGen Exome
Hyb Panel v2. Captures were performed using 100 ng Coriell DNA NA12878 in a singleplex reaction (n=3). Bioinformatics analysis was done for 3-way coverage correlation comparison, a representative pair from each vendor is demonstrated here. The IDT
xGen Exome Hyb Panel v2 shows a linear regression line that mimics the predicted one-to-one correlation line with an r2 value of 0.76. In this example, IDT outperformed the competitor.
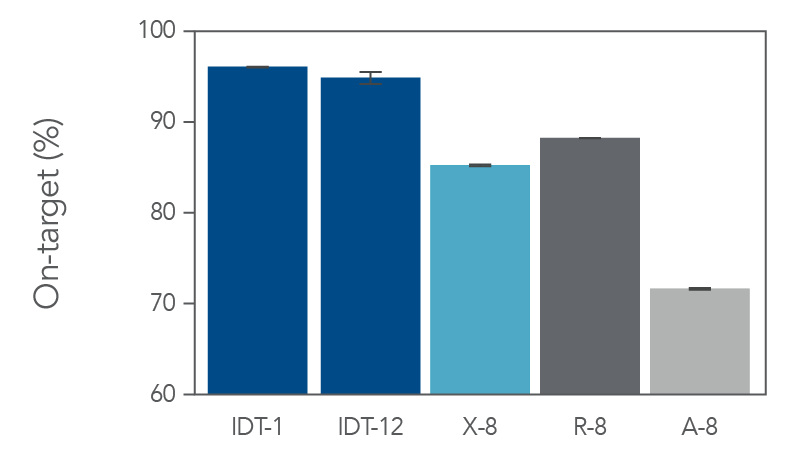
On-target coverage and uniformity of the xGen Exome Hyb Panel v2
Figure 2. The IDT whole exome sequencing workflow has higher on-target percentage in comparison to alternate suppliers. Sequencing libraries were prepared using enzymatic shearing and ligation-based library preparation using the library prep protocols from each vendor. Libraries were captured and multiplexed according to the vendor’s exome panel capture protocol (X-8, R-8, and A-8, where “8” signifies 8-plex captures, n=2). For IDT, we show both 1-plex (n=3) and 12-plex (n=2) captures (IDT-1 and IDT-12, respectively). Enriched libraries were sequenced on a NextSeq® instrument (Illumina) in high output mode using 2 x 100 bp paired-end reads for analysis. On-target bases were determined with Picard (percent selected bases) using 5 Gb per library.
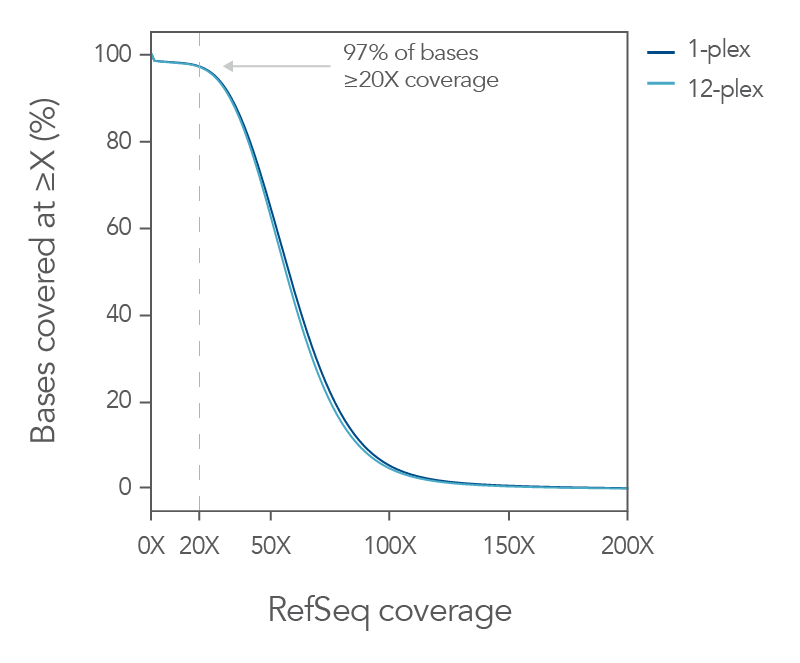
Figure 3. Consistent uniform sequence coverage with the xGen Exome Hyb Panel v2. DNA libraries were created from 100 ng of human genomic DNA (Coriell) and enriched either as 1-plex captures (n=3) or as a single
12-plex (n=2) capture using the xGen Exome Hyb Panel v2. The enriched libraries were sequenced (2 x 100 bp reads) on a NextSeq® instrument (Illumina) and subsampled to 5 Gb. The data shows uniform coverage with a flanked on-target rate
of 94.7%, mean target coverage of 64.5X, and a duplication rate of 3.3% (calculated with Picard).
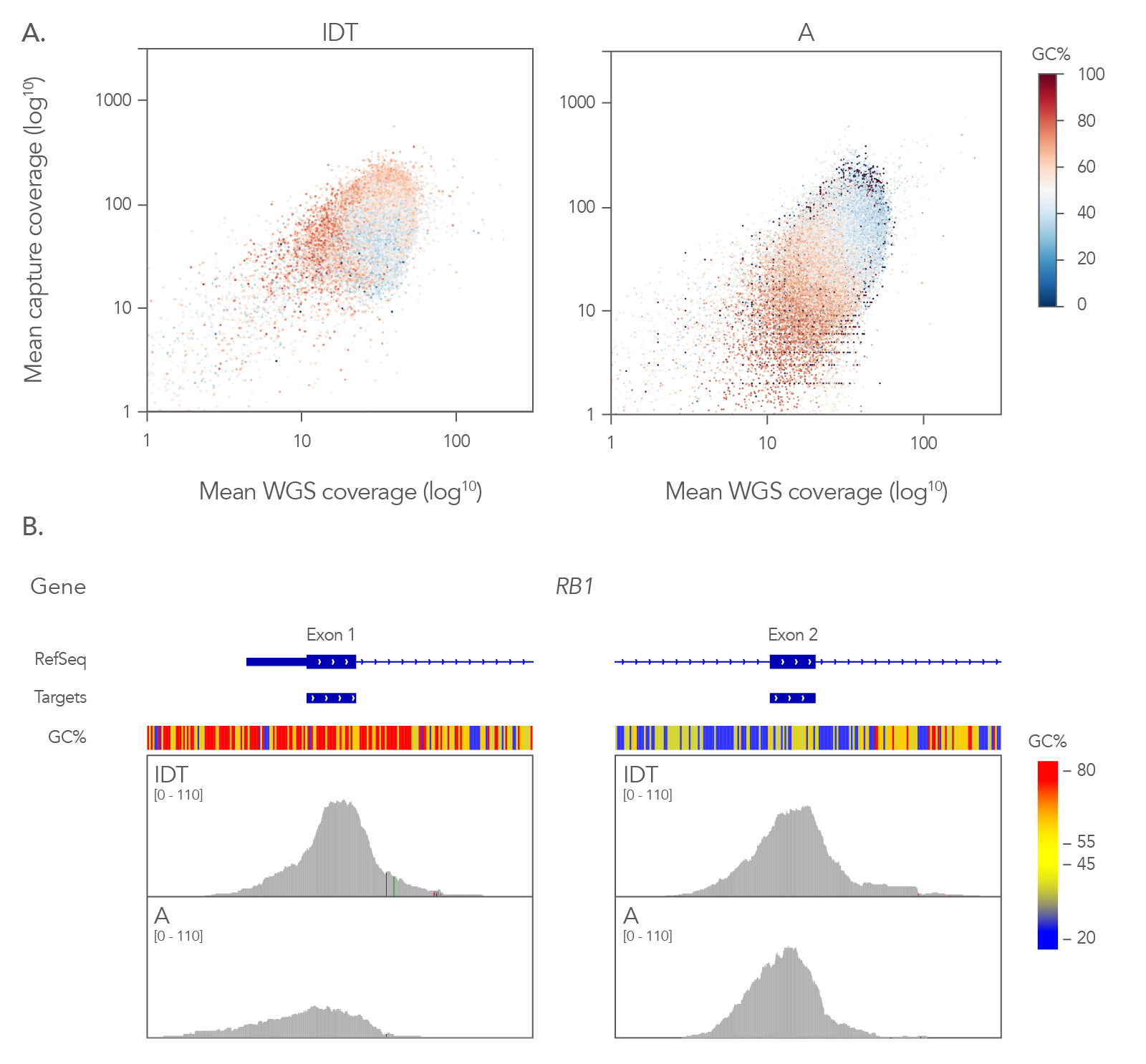
Coverage of RefSeq database with minimal sequencing
Figure 4. Coverage profile of the xGen Exome Hyb Panel v2 closely resembles whole genome data. (A) Comparing the coverage depths of different exome panels (8-plex, n=1) to coverage depth from whole genome sequencing (WGS) of a matched library shows that the xGen Exome Hyb Panel v2 closely matches WGS. Analysis of guanine-cytosine content (GC) shows that the xGen Exome Hyb Panel has a more effective capture of GC content (compare dark red and red dots in the right and left panels). (B) RB1 exons 1 and 2 show extremes of GC content with ~76% in exon 1 and ~38% in exon 2. A comparison of capture between IDT and supplier A shows a higher read depth across exon 1 for the xGen Exome Hyb Panel v2 on the Integrative Genomics Viewer (Broad Institute), whereas supplier A coverage has a lower number of reads of this exon with high GC content.
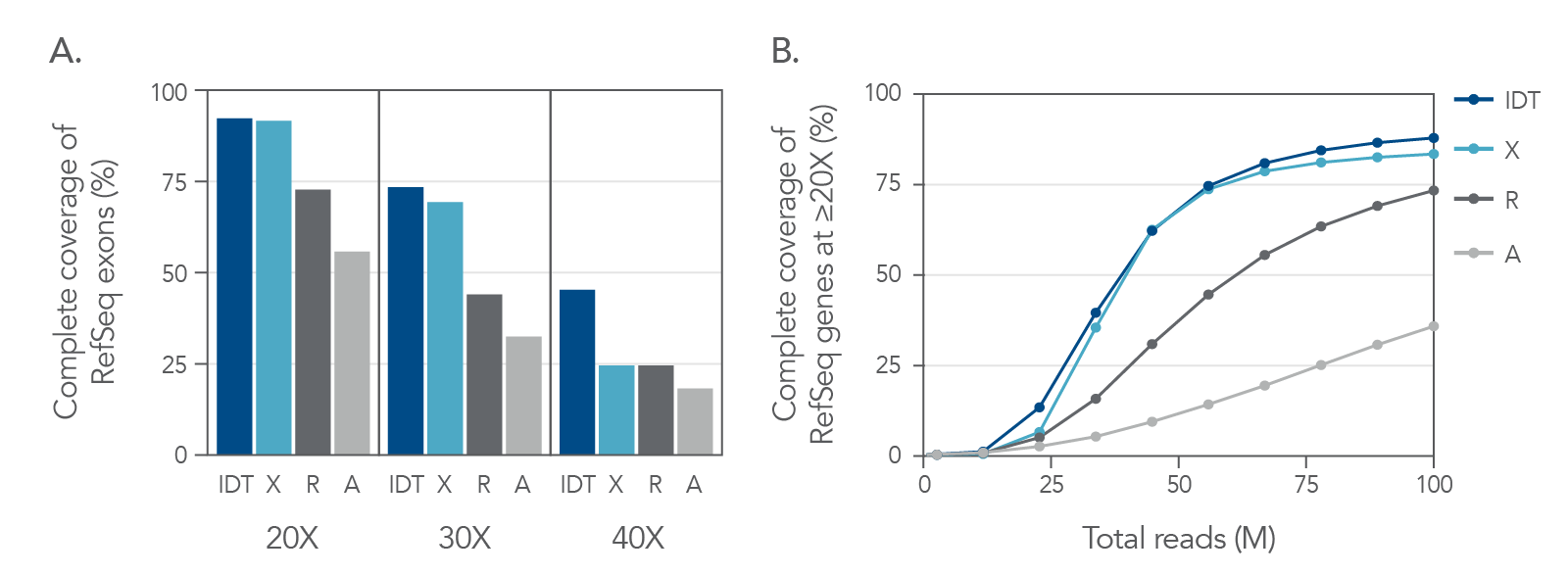
Figure 5. Exome coverage of the xGen Exome Hyb Panel v2. (A) 100 ng Coriell gDNA hybridization capture libraries (n=2) were sequenced with 5 Gb per sample while the percent of exons covered end-to-end at each read depth were calculated. The xGen Exome Hyb Panel v2 (IDT) shows the highest percentage of exons covered at each indicated depth (compared to suppliers X, R, and A). (B) The xGen Exome Hyb Panel v2 provides end-to-end RefSeq gene coverage at ≥20X. Individual samples were subsampled at different read depths (2 x 100 bp read length). The percentage of genes that were covered for every base of every exon at ≥20X was calculated at each read depth and plotted.
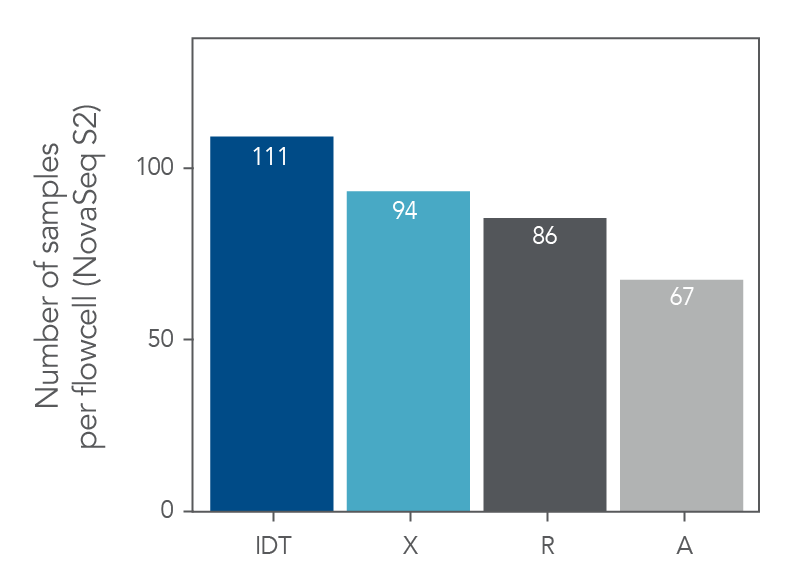
Maximize the number of samples per flowcell
Figure 6. xGen Exome Hyb Panel v2 can increase the number of samples loaded into a flowcell. DNA libraries were created from 100 ng of human genomic DNA (Coriell) and enriched either as 8-plex, n=1 (supplier R), 8-plex, n=2 (suppliers X and A) or 12-plex, n=2 (IDT) captures. The enriched libraries were sequenced (2 x 100 bp) on a NextSeq® instrument (Illumina) and the number of reads required to achieve 75X mean target coverage (Picard) per sample was estimated along with the number of samples that would fit on a NovaSeq™ S2 flowcell (Illumina).