Alt-R™ HDR Enhancer Protein
For researchers developing CRISPR-based therapies, this recombinant protein improves HDR efficiency across established and primary (i.e. iPSCs, and HSPCs ) cell types. Compatible with diverse Cas systems, available in RUO and soon CGMP options, it streamlines translational and clinical workflows and regulatory submissions.
Boost your edits, fast-track your breakthrough
Ordering
Alt-R™ HDR Enhancer Protein
- Excellent Performance: Consistently increases HDR rates up to 2-fold across diverse and challenging cell types like iPSCs, and HSPCs.
- Unmatched Precision: Enhances editing efficiency without increasing off-target effects, translocation rates, or compromising cell viability.
- Seamless Integration: Designed to fit effortlessly into your existing workflows, providing a smooth transition from discovery to the clinic with both RUO and CGMP (coming soon) options available.
- Accelerated Timelines: Boosts edited cell yields, shortening development cycles, and fast-tracking your therapeutic research programs.
- Trusted Quality: Manufactured by Aldevron™ under rigorous quality standards, ensuring consistency and reliability.
Alt-C™ HDR Enhancer Protein – Coming soon!
Designed specifically for CGMP/GMP therapeutic applications and manufactured by Aldevron to meet rigorous quality standards, Alt-C™ HDR Enhancer Protein integrates effortlessly into your existing workflows—empowering a smooth transition from research to clinical development. Be the first to know when it is available!
Get notifiedAssistant Professor of Pediatrics
Stanford University
This is an outstanding reagent with significant potential. Achieving 1.5x higher HDR efficiency while using less AAV (or potentially other DNA donor templates) is particularly valuable for therapeutic applications.
Product details
The Alt-R™ HDR Enhancer Protein is a recombinant ubiquitin variant engineered to increase the efficiency of homology-directed repair (HDR) during CRISPR-mediated genome editing. It functions by selectively inhibiting 53BP1, a key regulator that suppresses HDR by blocking end resection at double-strand break (DSB) sites (Bunting et al., 2010). By preventing 53BP1 recruitment, this protein-based enhancer shifts the DNA repair pathway balance away from non-homologous end joining (NHEJ) and toward HDR, enabling more precise genome modifications.
Alt-R HDR Enhancer Protein is optimized for RNP-based delivery during nucleofection, supporting efficient and direct incorporation into existing CRISPR workflows. The final engineered ubiquitin variant was identified through a high-affinity screen and was shown to improve HDR rates across multiple cell types and donor template formats, including single-stranded and double-stranded DNA. Critically, it achieves enhanced editing without the cytotoxicity or off-target effects sometimes observed with small-molecule inhibitors of the NHEJ pathway. Unlike DNA-PKcs inhibitors, the Alt-R HDR Enhancer Protein has minimal impact on NHEJ-mediated tag integration (e.g. in UNCOVERseq), preserving the utility of NHEJ where needed. Because it promotes a key step in the HDR pathway rather than directly inhibiting NHEJ, it can be used in combination with NHEJ inhibitors in workflows where such combinations are beneficial.
The Alt-R HDR Enhancer Protein is ideal for ex vivo genome editing applications where high editing efficiency and cell viability are essential, such as stem cell engineering for cell and gene therapy development (CGMP option coming soon).
Schematic overview of the main double-strand break (DSB) repair pathways following CRISPR-Cas nuclease activity. In the NHEJ pathway (left), repair is initiated by DNA-PK recruitment, followed by end processing and ligation, often resulting in indels. In the HDR pathway (right), the MRN complex initiates end resection, enabling coating of the resulting 3' ssDNA and template-directed repair using either double-stranded donors (Rad51-dependent) or single-stranded donors (Rad51-independent). The Alt-R™ HDR Enhancer Protein promotes HDR by inhibiting 53BP1, which otherwise blocks end resection, thereby enhancing knock-in efficiency without compromising cell viability or genomic integrity.
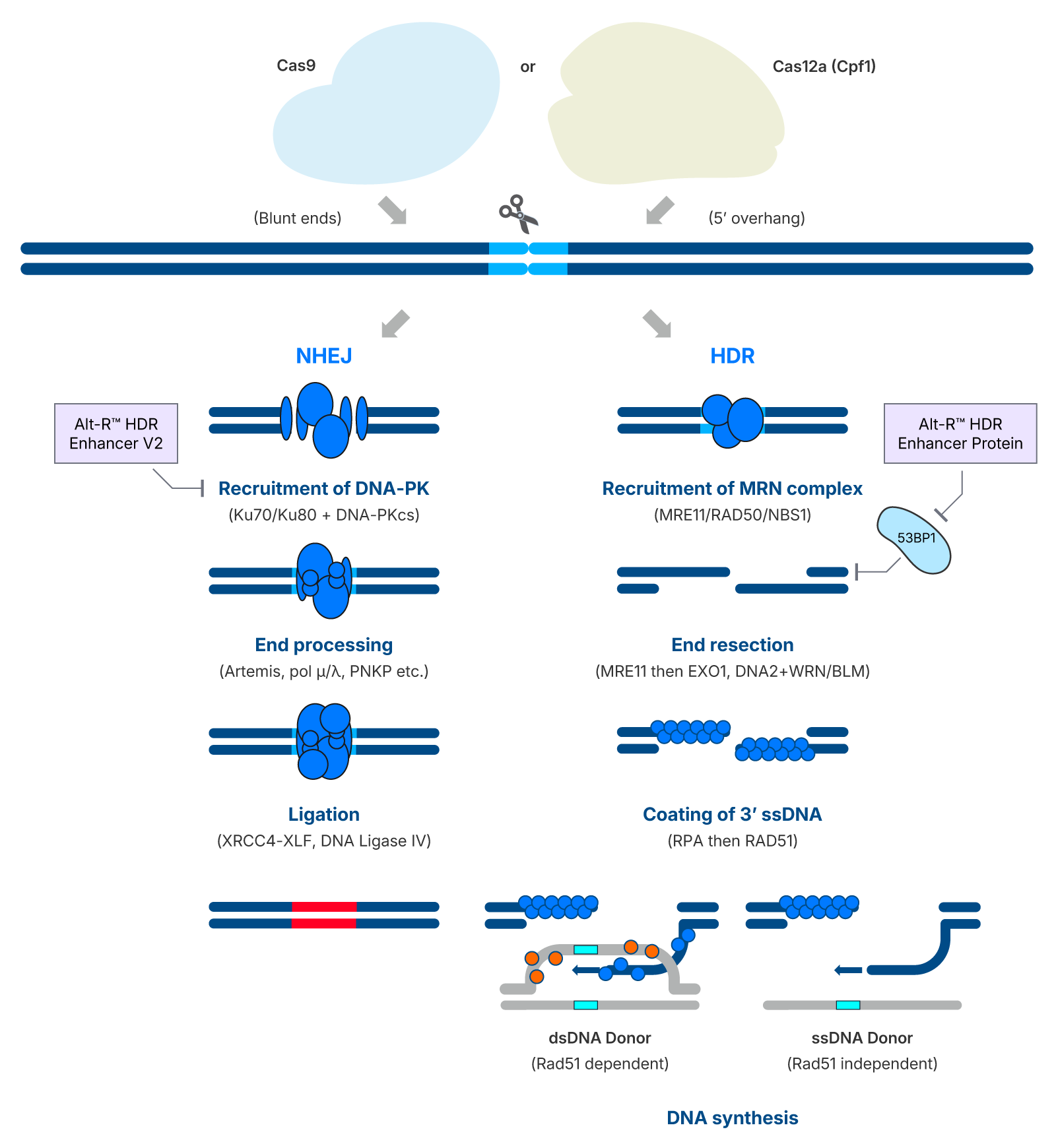
CRISPR-induced double-strand breaks are repaired via NHEJ or HDR. The Alt-R™ HDR Enhancer Protein promotes HDR by inhibiting 53BP1, making it specific to the HDR pathway, in contrast to DNA-PK inhibitors that enhance HDR by blocking the NHEJ pathway directly.
Product data
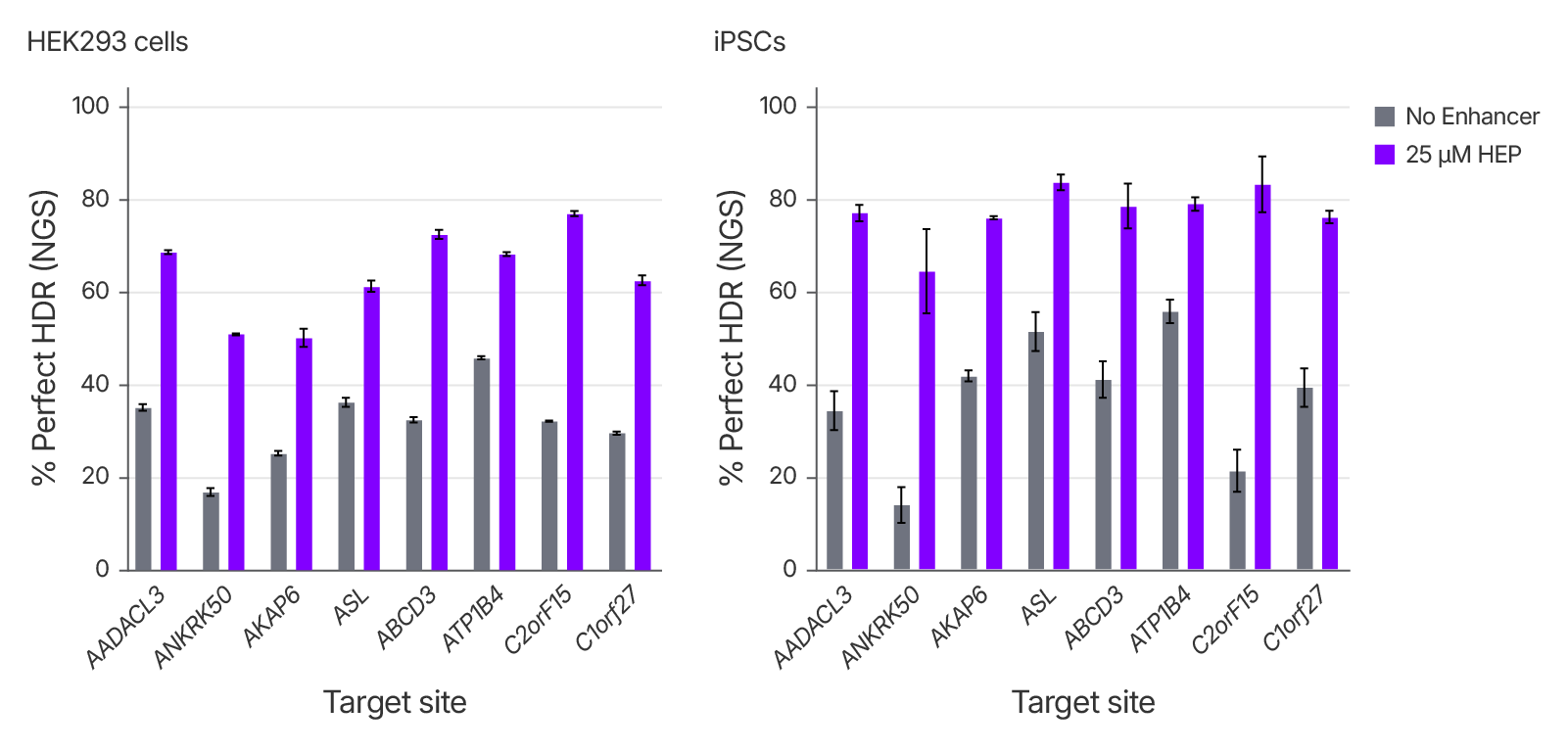
Alt-R™ HDR Enhancer Protein Increases HDR Efficiency across multiple loci and cell types
To evaluate the performance of Alt-R™ HDR Enhancer Protein on precise genome editing, we measured perfect HDR of a 6 bp insertion at eight genomic loci in two different cell lines, and showed consistent increase (up to 2-fold in different cell lines) in perfect HDR rates across all loci, including in primary iPSCs, highlighting the effectiveness of Alt-R™ HDR Enhancer in challenging-to-edit cell types.
Figure 1. Alt-R™ HDR Enhancer Protein (HEP) increases rates of HDR in multiple cell types. RNP complexes comprised of Cas9 and sgRNA targeting various loci were delivered to HEK293 cells at 2 μM or iPSCs (Coriell Institute) at 4 μM, with 2 μM ssDNA donor (Alt-R– HDR Donor Oligo), with or without Alt-R™ HDR Enhancer Protein using the 4D-Nucleofector System (Lonza). HDR was assessed using NGS. Error bars indicate SD, n = 3.
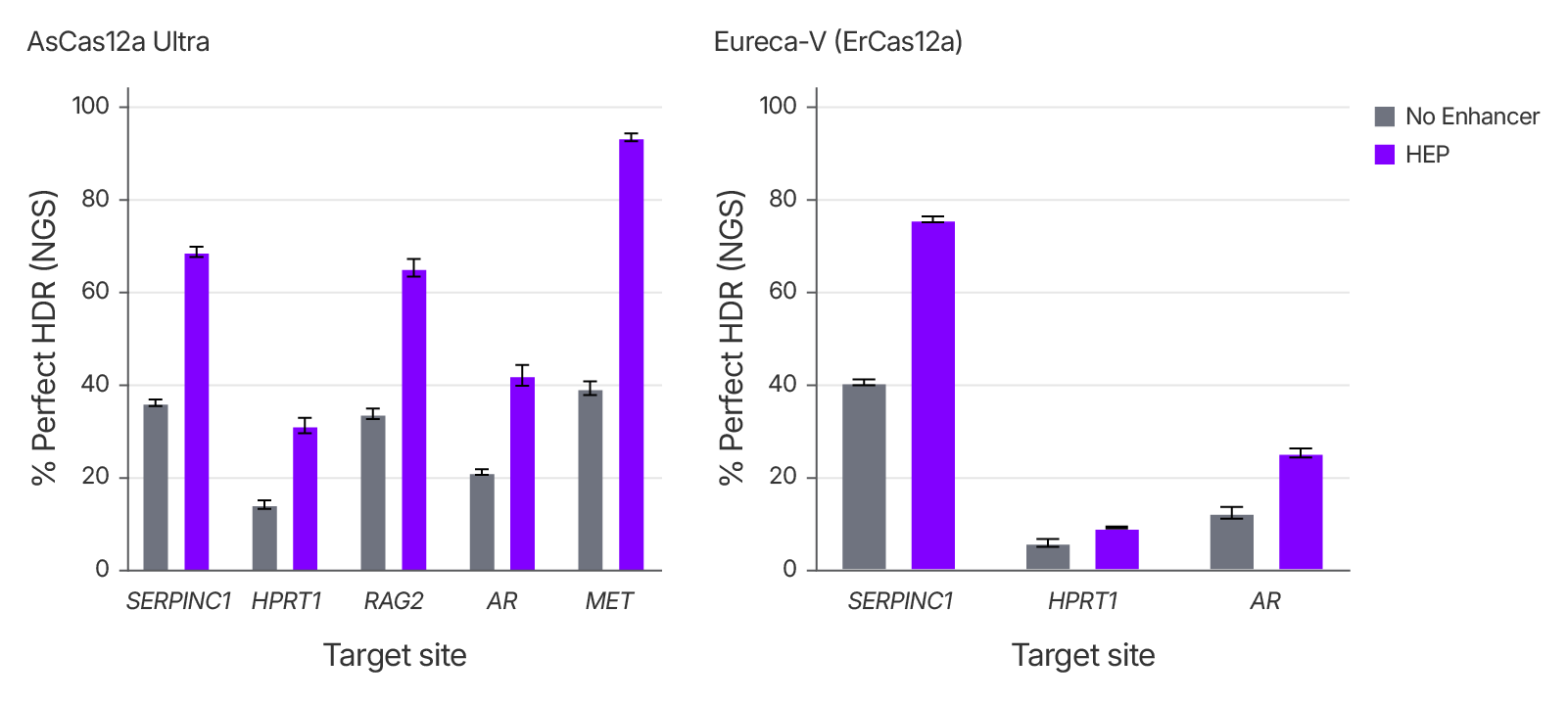
Alt-R™ HDR Enhancer Protein increases HDR efficiency with multiple CRISPR nucleases
To evaluate the compatibility of Alt-R™ HDR Enhancer Protein with different CRISPR nucleases, we tested its effect on perfect HDR efficiency using Alt-R™ A.s. Cas12a (Cpf1) Ultra and Aldevron Eureca-V™ in HEK293 cells (see Fig. 1 for results obtained with Cas9). The Alt-R™ HDR Enhancer Protein significantly improved HDR efficiency across various target sites, demonstrating its broad utility across diverse CRISPR systems, including those generating staggered DNA breaks.
Figure 2. Alt-R™ HDR Enhancer Protein increases rates of HDR with nucleases that produce staggered cuts. RNP complexes comprising crRNAs targeting various loci and Alt-R™ A.s. Cas12a (2 μM) or Eureca-V™ (3 μM), with 2 μM Alt-R HDR Donor Oligo and either 0 or 25 µM HDR Enhancer Protein (HEP) were delivered to HEK293 cells using the 4D-Nucleofector System (Lonza). Genomic DNA was extracted after 48 hours and HDR was assessed by NGS on a MiSeq® platform (Illumina®) using 2 x 150 PE sequencing.
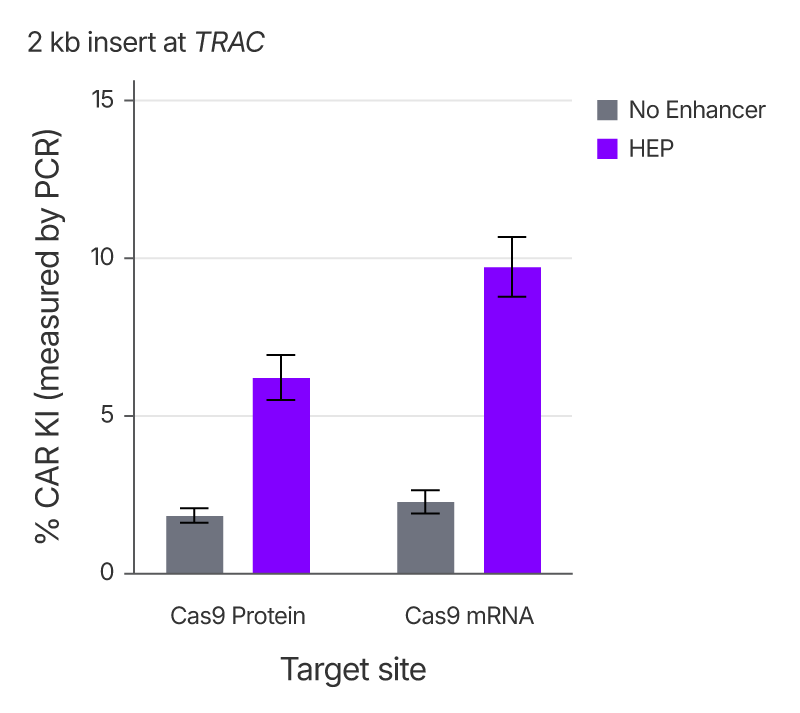
Alt-R™ HDR Enhancer Protein improves knock-in efficiency across Cas9 delivery formats
To evaluate the compatibility of HDR Enhancer Protein with different Cas9 delivery formats, we compared knock-in efficiency of an approximately 2 kb chimeric antigen receptor (CAR) sequence at the TRAC locus in HEK293 cells using either Cas9 mRNA or Cas9 RNP. In both formats, the addition of the enhancer resulted in a substantial increase in the knock-in rates, demonstrating its flexibility in diverse editing workflows.
Figure 3. Alt-R™ HDR Enhancer Protein boosts HDR when paired with either Cas9 protein or Cas9 mRNA. HEK293 cells were treated with either 2 µM RNP or 1 µg Cas9 mRNA plus 4.8 µM sgRNA, 1 µg Alt-R™ HDR donor block, and either 0 or 25 µM Alt-R HDR Enhancer Protein using the 4D-Nucleofector System (Lonza). CAR knock-in efficiency was calculated using PCR followed by analysis with an Agilent Fragment Analyzer and PROSize 3.0 software. Error bars indicate SD, n = 3.
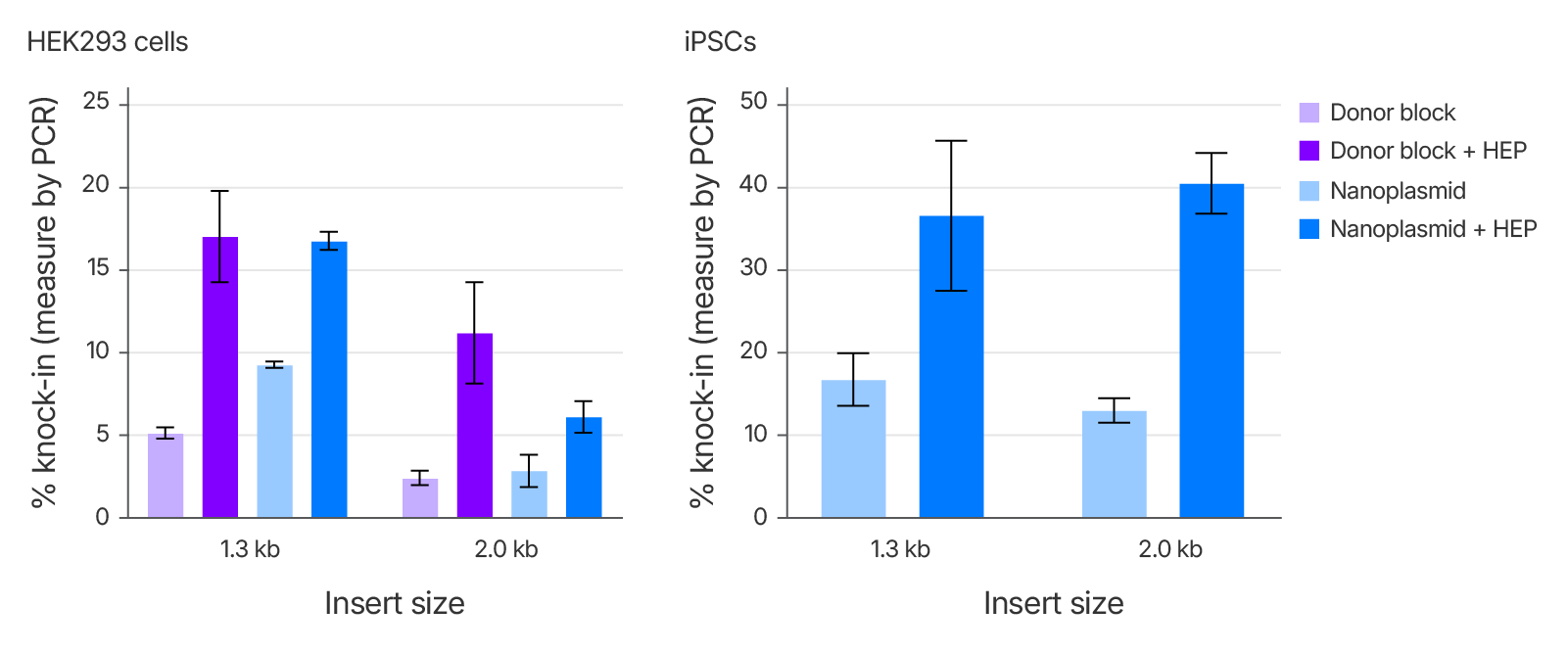
Alt-R™ HDR Enhancer Protein improves large knock-in efficiency using different donor types
To test compatibility with different types of large HDR templates, we performed knock-in at the TRAC locus of 1.3–2.0 kb CAR sequences in either Alt-R™ HDR Donor Blocks or Aldevron Nanoplasmid vectors in HEK293 cells and iPSCs. The presence of Alt-R™ HDR Enhancer Protein significantly increased integration efficiency, underscoring its effectiveness in supporting large genomic insertions with multiple donor types and across cell types.
Figure 4. Alt-R™ HDR Enhancer Protein is effective when used with multiple types of donor template. Cas9 mRNA and TRAC sgRNA were delivered with either an Alt-R HDR Donor Block or a Nanoplasmid with 500 bp homology arms and either 0 or 25 μM Alt-R HDR Enhancer Protein using the 4D-Nucleofector System (Lonza). HEK293 cells were treated with 1 µg Cas9 mRNA, 4.8 µM TRAC sgRNA, and 1 µg of donor. iPSCs were treated with 2 µg Cas9 mRNA, 5 µM TRAC sgRNA, and 2 µg of Nanoplasmid. Knock-in efficiency was calculated using PCR and analysis with PROSize 3.0 software (mean ± SD, n = 3).
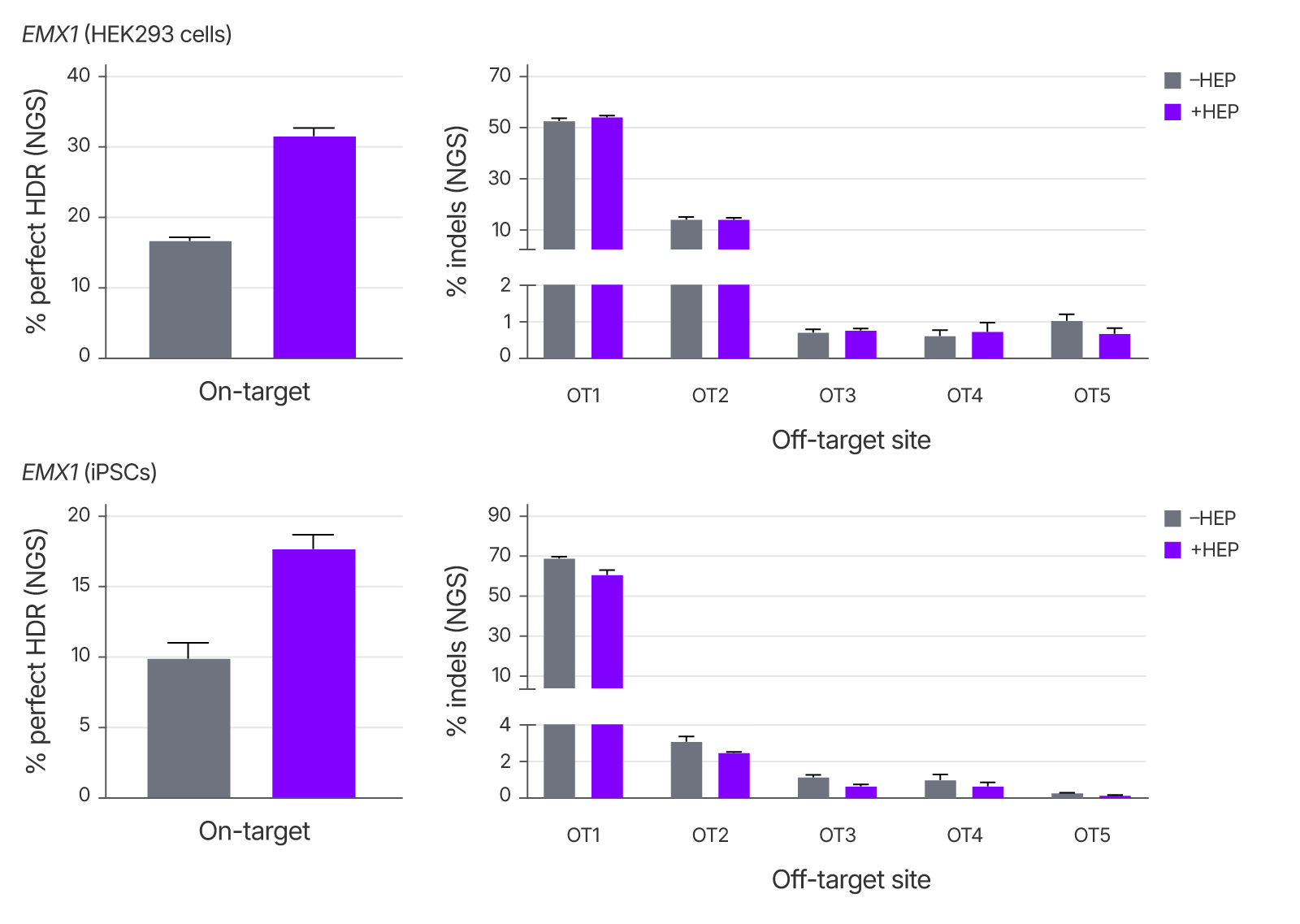
Alt-R™ HDR Enhancer Protein improves on-target editing without increasing off-target indels.
To determine the specificity of genome editing in the presence of Alt-R HDR Enhancer Protein, we measured both on-target insertion of 6 bp by HDR and off-target indel formation following Cas9 RNP delivery targeting the EMX1 locus. The addition of HDR Enhancer Protein improved on-target HDR rates while maintaining off-target indel frequencies in both HEK293 and iPSC cells, supporting its use in high-fidelity editing applications.
Figure 5. Use of Alt-R™ HDR Enhancer Protein does not increase indels at off-target sites. 2 μM RNP comprising Cas9 and sgRNA targeting EMX1, 2 μM ssDNA Alt-R HDR Donor Oligo, and 0 or 25 HEP (HEK293 cells) or 12.5 µM uM HEP (iPSCs) were delivered using the 4D-Nucleofector System (Lonza). Editing was measured after 48 hours using rhAmpSeq™ NGS targeting the EMX1 on-target site and a set of known off-target sites. Error bars indicate SD, n = 3.
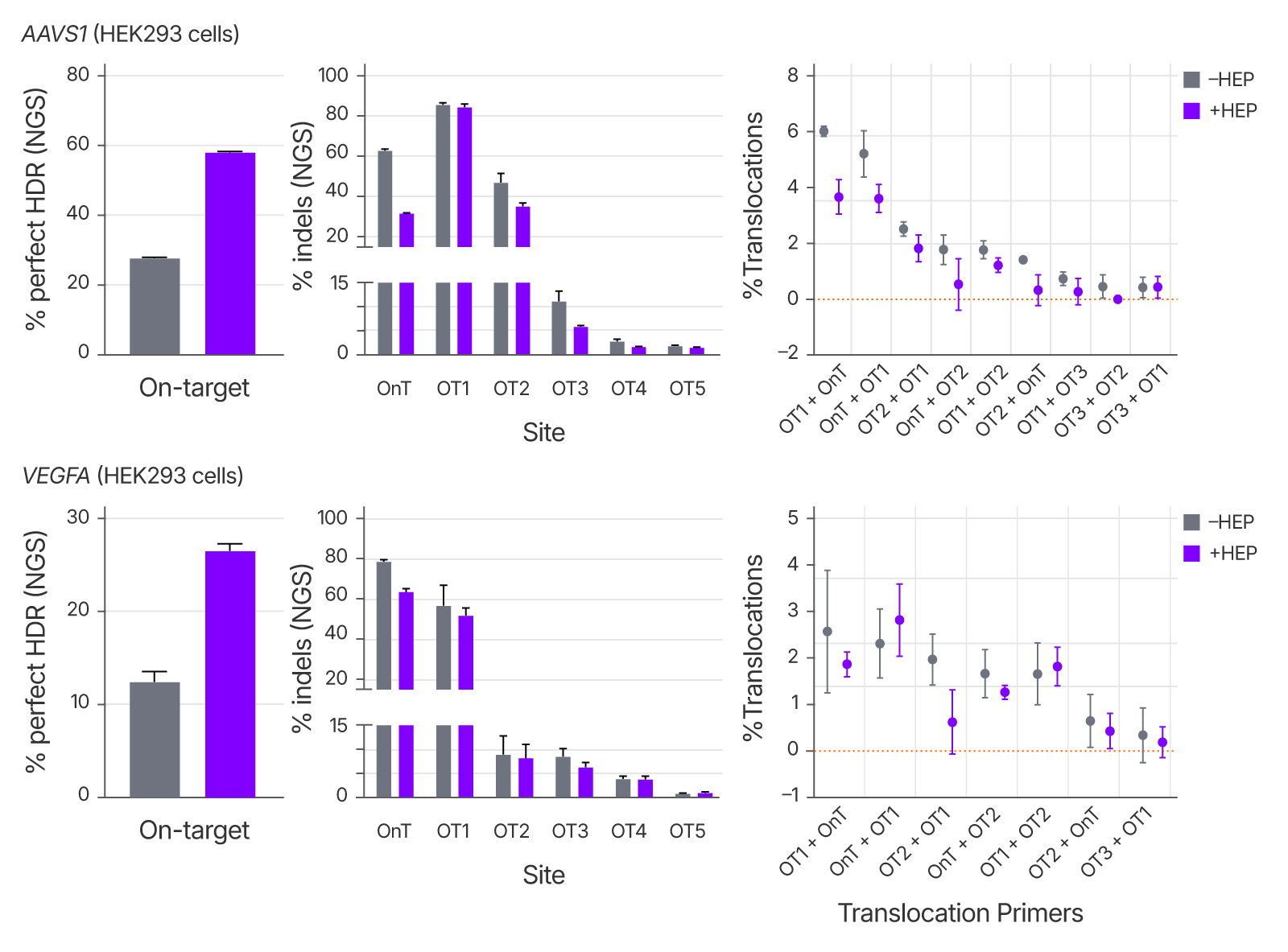
Alt-R™ HDR Enhancer Protein boosts HDR without increasing translocation frequency.
Genotoxicity was evaluated by quantifying chromosomal translocation events between known cut sites using sgRNAs with high off-target risk (e.g., AAVS1 and VEGFA). Alt-R™ HDR Enhancer Protein increased HDR rates at the target site without elevating translocation frequency, confirming its suitability for applications requiring both precision and genomic stability.
Figure 6. Use of Alt-R™ HDR Enhancer Protein does not increase translocations between known Cas9 cut sites. 2 µM Cas9 RNP targeting AAVS1 or VEGFA, 2 µM Alt-R HDR Donor Oligo, and either 0 µM or 25 µM Alt-R™ HDR Enhancer Protein (HEP) were delivered to HEK293 cells using the 4D-Nucleofector System (Lonza). Genomic DNA was extracted after 48 hours. (Left) On-target HDR and off-target indels were measured by NGS. (Right) Translocations were quantified using Primer Anchored Statistical Translocation Analysis (PASTA, Kinney et al., 2025.), in which mismatched primer pairs spanning known RNP cut sites (x-axis) were identified in NGS reads. Only translocations meeting stringent criteria and detected in at least 2 of 6 replicates are shown. Error bars indicate SD, n = 3 per condition.
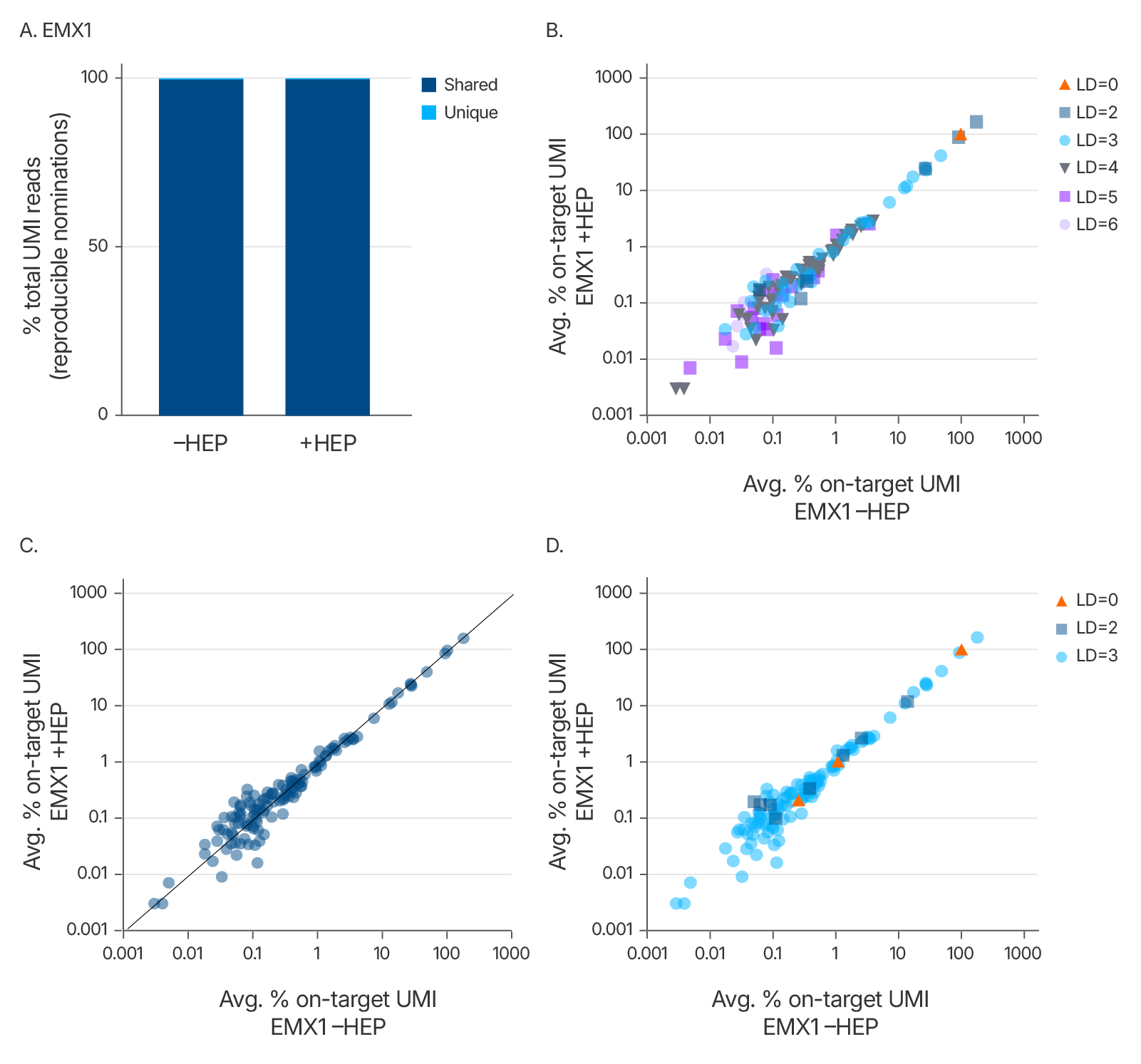
Alt-R™ HDR Enhancer Protein does not introduce new Cas9 off-target sites
To assess the potential for new off-target sites, we used UNCOVERseq to profile off-targets in HEK293 cells stably expressing Cas9. The Alt-R™ HDR Enhancer Protein did not create new off-target sites and showed off-target profiles consistent with no HDR Enhancer Protein, confirming its compatibility with high-specificity genome editing.
Figure 7. Alt-R™ HDR Enhancer Protein does not increase Cas9 off-target sites. 5 µM sgRNA targeting EMX1, 0.5 µM dsODN tag, and either 0 or 50 µM Alt-R HDR Enhancer Protein were delivered to HEK293-Cas9 cells (CRL-1573) using the 4D-Nucleofector System (Lonza). UNCOVERseq was used to nominate off-target sites 72 hours post-treatment, as described in Kinney et al., 2025. (A) Percent of total unique molecular identifier (UMI) reads at reproducible sites (Levenshtein distance <7, found in ≥2 samples) that are unique to or shared between –HEP or +HEP conditions. (B–D) Scatterplots of the average % off-target UMI reads relative to on-target UMI reads for each off-target site shared between –HEP and +HEP conditions, categorized by (B) Levenshtein distance, (C) correlation, and (D) Tier classification. The Tier system reflects predicted biological impact and annotation: Tier 1: High MAR score and exon annotation or Levenshtein distance = 0, Tier 2: Consensus in ≥2 replicates and exon annotation, Tier 3: Exon annotation, high MAR score, >1% UMI reads, or consensus in ≥2 replicates.
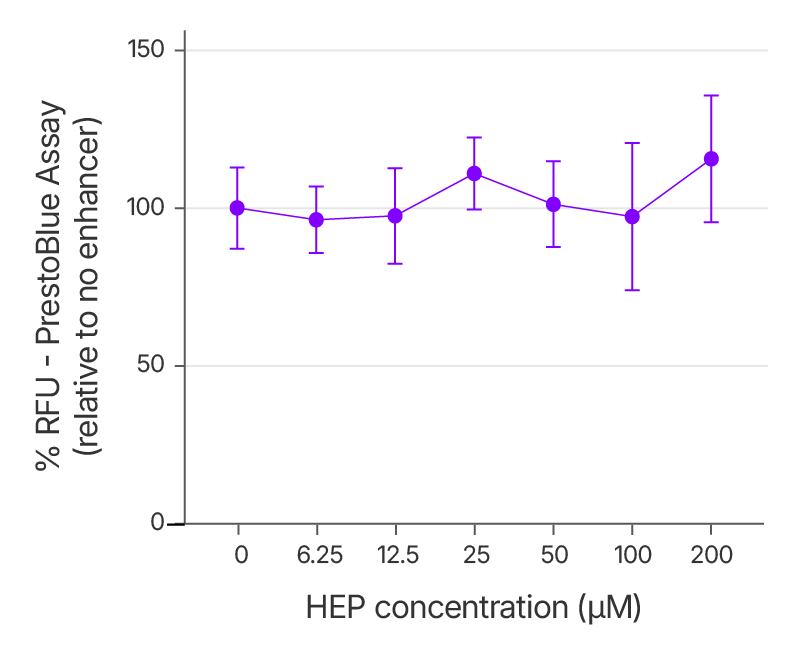
Alt-R™ HDR Enhancer Protein does not induce cellular toxicity across a wide concentration range
To evaluate the cytotoxicity profile of the Alt-R™ HDR Enhancer Protein, we assessed cell viability in HEK293 cells with increasing concentrations of the enhancer. Results from a PrestoBlue assay show that even at elevated doses, Alt-R™ HDR Enhancer Protein does not reduce cell viability, confirming its suitability for use in sensitive cell types and high-efficiency editing protocols.
Figure 8. No cytotoxicity observed with increasing doses of Alt-R™ HDR Enhancer Protein. 2 μM RNP complex targeting EMX1, 2 μM ssDNA Alt-R HDR Donor Oligo, and varying concentrations of Alt-R HDR Enhancer Protein were delivered to HEK293 cells using the 4D-Nucleofector System (Lonza). Cell viability was measured 48 hours post-treatment using the PrestoBlue assay, with results reported as percent relative fluorescence units (RFU) compared to untreated controls. Error bars represent standard deviation (SD); n = 3 biological replicates.
Resources
Frequently asked questions
How do I optimize the dose of the Alt-R HDR Enhancer Protein?
Does the Alt-R HDR Enhancer Protein increase rates of large KI?
What cell types and donor types does the Alt-R HDR Enhancer Protein work best with?
The Alt-R HDR Enhancer Protein has been shown to increase HDR rates in a variety of cell types (U2OS cells, Jurkat cells, HEK 293 cells, iPSCs, HSPCs, and T cells) and using a variety of donor types including ssDNA, linear dsDNA, plasmid DNA, and AAV template. The level of increase is often dependent on the combination of the cell line and donor type. For example, while the Alt-R HDR Enhancer Protein has been shown to boost HDR rates in T cells with non-viral donors and in HSPCs with an AAV donor, it has low effectiveness when used in T cells with an AAV donor. We recommend testing a range of doses with your cell line and donor to determine the level of effectiveness of the Alt-R HDR Enhancer Protein in your specific application.
Does the Alt-R HDR Enhancer Protein increase rates of HDR with other CRISPR Cas enzymes besides Cas9?
Does the Alt-R HDR Enhancer Protein increase off-target editing or translocation events?
Can I use the Alt-R HDR Enhancer Protein and the Alt-R HDR Enhancer V2 in combination to increase HDR rates even more?
Both Alt-R HDR Enhancers can be used in the same experiment and can have an additive effect on rates of HDR. The two enhancers are delivered to cells differently and at different steps during CRISPR editing. Though the Alt-R HDR Enhancer Protein often results in a smaller increase in HDR than the Alt-R HDR Enhancer V2 in some cell lines, the Alt-R HDR Enhancer Protein has a better off-target and toxicity profile and the CGMP version will be available soon to address the critical need for clinical applications.
Learn more: Alt-R™ HDR Enhancer Protein: Improving homology-directed repair from bench to bedside
What's the difference between the Alt-R HDR Enhancer Protein and the Alt-R HDR Enhancer V2?
The two HDR enhancers target different cellular DNA repair pathways and increase rates of HDR by different mechanisms. The Alt-R HDR Enhancer Protein inhibits 53BP1 to promote DNA break end-resection, a critical step in cellular HDR. The Alt-R HDR Enhancer V2 is a small molecule that inhibits the NHEJ pathway to indirectly bias dsDNA break repair toward HDR.
Related products
References
- Kinney KJ, Jia K, Zhang H, et al. UNCOVERseq Enables Sensitive and Controlled Gene Editing Off-Target Nomination Across CRISPR-Cas Modalities and Systems.bioRxiv (Cold Spring Harbor Laboratory). Published online May 11, 2025. doi:https://doi.org/10.1101/2025.05.09.653165
- Bunting SF, Callén E, Wong N, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks.Cell. 2010;141(2):243-254. doi:10.1016/j.cell.2010.03.012